Programming in a research laboratory
I started my scientific work experience in a lab that studies brain diseases using MRI and other modalities. I then spent sometime in industry learning to work at a faster pace and with newer technology.
Now I’m back in research and I’m finding myself using a whole new set tools and principals.
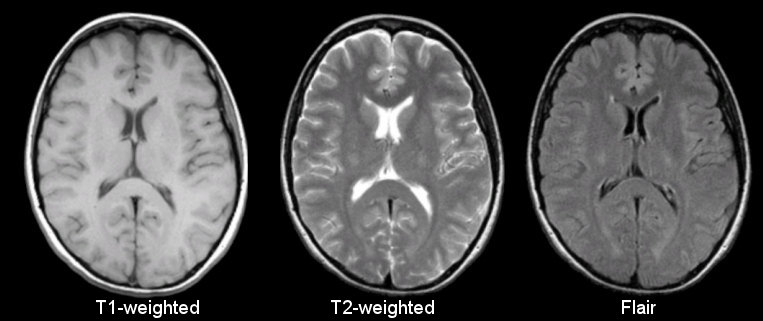
I’m going to focus on organizing inputs to processing work flows, which in my case are brain MRIs. Typically a study will have subjects, which undergo sessions, that have runs. The MRI Scanners produce files in the medical format known as DICOM. The meta data stored as date-times in the file name and some DICOM header tags.
The files are brought into a researchers file system in a flat format. Then someone puts them ins some order that makes sense to humans.
- Subject
- Session
- Modality
- runs
- Modality
- Session
Why not just keep the files where they are ?
I’ve seen a lab where symbolic links where used to organize the data without having to copy it around. That was highly annoying when it came time to rsync the data to my lab’s storage. The files had been organized into the above hierarchy already, but since I didn’t have all the links setup I couldn’t preserve the structure.
I could just leave the DICOMs in place and use a database.It’s easier to wrangle the meta data via SQL and Python/Pandas than to write file walking scripts.
Post Doc Programmers
The worst kind of software I’ve been handed are MATLAB scripts which build paths to data inputs. I’ve had to work with more than one complex, monolithic program that required a complex and opaque data structure that itself required a complex and opaque script to get working.
Often times the researchers are unclear what data they, where it is located and if it’s homogeneous or not. Especially with research tools which do not typically enforce a standard just merely allow it, or not.
A Better Approach: A Data Base
The most organized lab I worked in and where I got my start, used a well thought out system to store meta data in a SQL database. This database was curated and maintained carefully. It was the life line of the lab which processed and stored several terabyte of imaging data for research.
I also learned during my short stint as a consultant, that generating an invoice for goods delivered and received is pretty crucial. When given a data set, it’s wise to inventory it and send a receipt for what was given. Especially if you suspect there will be issues regarding the quality or meta data inconsistencies.
I have written a handful of inventory programs which work with any type or group of files and a few particularly for medical imaging storage, e.g. DICOM and NiFTI1. Even if I have to start with a legacy file system I can create a DB.
The first thing you’ll want to do is to rebuild some of the structure that was lost going from a directory structure to a flat inventory file.
Here’s some steps that I might take to organize data that I want to import:
SELECT COUNT(`fullpath`),
CASE
WHEN `fullpath` LIKE '%/subA/%' THEN `SubA`
WHEN `fullpath` LIKE '%/subB/%' THEN `SubB`
ELSE `Neither` END AS `Sub`
FROM `inventory`
GROUP BY `Sub`Screen shot of SqliteStudio.
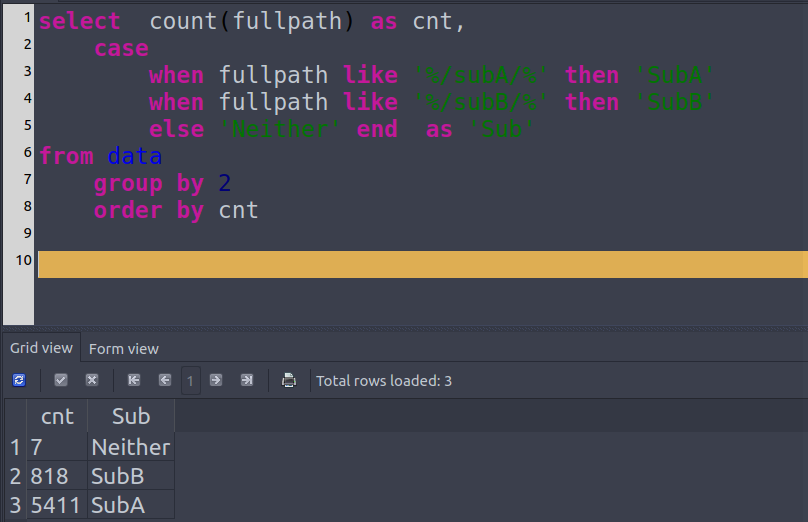
I typically prefer to make a categorical column to repeat this procedure easily since it’s a simple and necessary sanity check.
ALTER TABLE `inventory` ADD COLUMN `Sub`
UPDATE TABLE inventory
VALUES = CASE
WHEN `fullpath` LIKE '%/subA/%' THEN `SubA`
WHEN `fullpath` LIKE '%/subB/%' THEN `SubB`
ELSE `Neither` END AS `Sub`
FROM `inventory`Now our basic sanity check is easier.
SELECT COUNT(`fullpath`) AS CNT, `Sub`
FROM `inventory`
GROUP BY `Sub`
ORDER BY CNTShort Cuts to Creating Smallish DB’s
It can be a pain to make a DB for a legacy data set or just to practice. The inventory program I mentioned above can be sped of significantly using GNU Parallel.
One is to use CSV-KIT which uses Python to provide nice command line programs. My prefered method,is to use Pandas to create a data frame, which will right away help debug your inventory csv generation as it will fail to load if the rows are not homogeneous. I’ve read a 3.3 million row csv file into Pandas in about 30 seconds. Not lightning fast but totally useable.
Then using Sqlalchemy to connect to SQLITE and finally push the data frame to a SQLITE database file.
ipython import pandas as pd from sqlalchemy import
create_engine
df = pd.read_csv("inventory_file.csv") # if a line fails the exception will
be specific about which line which is great engine=
create_engine("sqlite:////path/to/inventory_database.db")
df.to_sql("inventory", engine)SQLITE
I mentioned SQLITE above. I’m somewhat new to SQL and databases and I’ve found that SQLITE3 is very easy to get going. The DB is a file that can be shared with co-workers. No need to setup a MySQL for prototyping. UPDATE: Using Docker, I’ve found it’s very quick to get a MySQL instance running in a VM. But I had to wait for IT to setup the VM.
I like SqliteStudio as GUI client for SQLITE.(http://sqlitestudio.pl)[SqliteStudio] It’s not in the Debian or RHEL repos, but it is a standalone executable so there’s no installation, but untar it and run.
Conclusion
So far no one has complained about this system. They are learning a few SQL commands to select their data. Overall it’s a lot fewer lines of code than the MATLAB they would write.
Using this system, sanity checking is built into the processing of receiving data.
